Understanding Bison-Cattle Introgression
In 1887, William T. Hornady seeing what ranchers were doing and all the cross-breeding of bison and cattle wrote "…it is to be feared that it will soon become a difficult matter to find a buffalo of absolutely pure breed."
"There is reason to fear that unless the United States Government takes the matter in hand and makes a special effort to prevent it, the pure-blood bison will be lost irretrievably through mixture with domestic breeds and through in-and-in breeding." William T. Hornaday, The Extermination of the American Bison, 1889.
Introgression is the transfer of genetic material from one species into the gene pool of another by the repeated backcrossing of an interspecific hybrid with one of its parent species. In very simple terms, it is the breeding between bison and domestic cattle and then breeding back the hybrid to one of the original species.

Due to back-breeding, bison hybrids (bison with evidence of cattle introgression) are usually indistinguishable from pure bison.
Beginning as far back as the 1700's there are records of bison being crossed with cattle, either accidentally or on purpose. However, it was not until 1995 that Polzeihn and coworkers (1), while constructing the phylogeny of North American bison in the Custer State Park herd of South Dakota, inadvertently discovered cattle-type mitochondrial DNA within the bison population. Since that time there have many scientific articles attesting to the level of bison-cattle introgression throughout the bison populations (2,3).
Most of the introgression can be traced back to the late 1800's to early1900's when most of the ranchers of the so called "bison foundation herds" from which all modern day bison have their origins, experimented with bison cattle crosses. These included Charles Goodnight, Michael Pablo, Charles Allard, Charles "Buffalo" Jones, Scotty Phillip, and others (4-8). They called these hybrids "cattalo" and it was an attempt to integrate some of the bison traits into domestic cattle. They assumed these hybrids would display heterosis (hybrid vigor or outbreeding enhancement) whereby the offspring would be superior to the parents.
These ranching experiments to crossbreed bison and cattle did not produce the results they wanted or anticipated and, as commonly occurring when crossing distantly related animals, there was outbreeding depression (a reduction in fitness) rather than hybrid vigor. As a result, these early experiments were aborted as unsuccessful (although for some reason they continue by a few to this date). See our article entitled "Bison Hybrids, Cattalo, and Beefalo".
Since the discovery of cattle introgression in bison in 1995, there have been no efforts to preserve what may be left of the pure bison population. By disinformation, misinformation, general misunderstandings, and maybe even some dishonesty, cattle genes have continued to be spread within the bison population. Even the Bison Registry of the National Bison Association (NBA) requires genetic testing but allows bison to be registered with up to 3 distinct cattle alleles (introgression markers) as long as they look like bison. And the presence of cattle introgression within these registered bison is considered "proprietary" (hidden) information by the NBA and is not publically available.
There have always been 2 sides of thought on the issue. There are the purists that think the original genome of the bison needs to be preserved and the other side, primarily meat producers, which claim what does it matter? There are valid arguments on both sides:
- For the meat producer, who cares? Is there a difference in the meat between a pure bred Angus cow and an Angus cross?
- For the purists, there is validity in maintaining the pure wild bison genes and limiting human selection interference (hybridization). Is there a reason and value to saving any species?

Not all bison hybrids are this obvious. White-yellow bison are signs of the Charolais SILV dilution gene.
Initial testing for cattle introgression only involved the examination of mitochondrial DNA (mtDNA) which is maternally derived (passed to offspring by the mother). This test had limited value since a cattle bull crossed with a bison cow (50% hybrid) would be categorized as pure bison since the offspring would inherit the bison cow mtDNA. Later, nuclear microsatellite markers or alleles (tracts of repetitive DNA often called short tandem repeats) were incorporated in standard testing to detect nuclear (maternal and paternal) cattle DNA. Initially 14 microsatellite markers were used which was expanded to 25 in later years although the 14 original continued to be used by some diagnostic centers.
Based on mtDNA and the original 14 nuclear introgression markers, a number of herds were found to be free of cattle introgression. Although most private and public bison herds were found to contain cattle genes and evidence of introgression, there were a few herds found to be free of cattle genes. These included the Yellowstone, Wind Cave, Henry Mountain, and Elk Island herds (3). Most private and other Federal herds tested were found to be genetically polluted with cattle genes to various degrees (2,3).
A study in feedlot bison suggested that the presence of cattle mtDNA in bison resulted in a significant difference in weight gains and size between bison with bison mtDNA compared with bison with cattle mtDNA (9). This study ignored nuclear evidence of introgression and led to the conclusion by many that only mtDNA was important. For obvious reasons, this conclusion is seriously flawed. Nonetheless, the National Bison Registry allows "registered" bison to contain nuclear microsatellite markers of introgression as long as the bison did not contain cattle mtDNA. It also led to the claims that bison are free of cattle introgression and cattle genes based solely on mtDNA. This erroneous reliance on mtDNA continues to this date and allows the National Bison Association to mistakenly claim that less than 6% of bison have evidence of cattle introgression and has promoted the continued pollution of the bison genome.
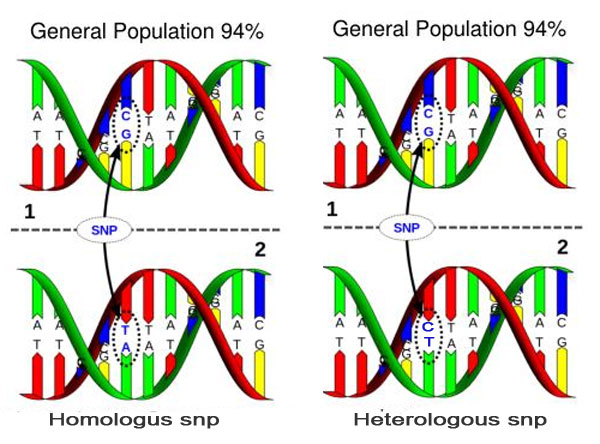
Figure 1. SNPs may be a complete nucleotide switch (homologous) or a simple mismatch (heterologous).
Whole genome single nucleotide polymorphism (SNP, pronounced "snip") genotyping is one the latest and state-of-the-art methodology used for many genetic studies, including introgression research (10). There are only 4 nucleotides or nucleobases that make up DNA: guanine (G), adenine (A), cytosine (C), and thymine (T) that match in the double helix as A-T and G-C. A substitution of the normal pair within a sequence is called a SNP, e.g., rather than G-C (normal), it is T-A. There may also be heterologous mismatches such as G-A, G-T, C-T, C-A. (Figure 1). These snp's may do nothing or they may alter the function of a gene by inactivating or activating the gene. To learn more about SNPs see Making SNPs Make Sense.
In 2020, a joint venture between the Canadian Bison Association (CBA) and NeoGen Corpdeveloped an introgression diagnostic test looking at 10,000 putative introgression markers identified by whole genome snp genotyping. The development of this test revealed introgression as high as 4.5% in their 461 validation animals (personal communication) and identified hybridization in a large number of bison including those previously tested clean based on mtDNA and nuclear satellite markers, attesting to the superior application of this test to mtDNA and nuclear satellite markers. The test also identified some individual animals that were allegedly free of cattle introgression. Although the development of this test has never been published within the scientific community, the methodology used for cattle introgression analysis are said to be the same as those used in their sub-species composition test (11). This bison introgression test is currently available through the Canadian Bison Association. Since this study has never been published or subject to critical peer review, the validity and reliability of this test must be questioned. Nonetheless, it is a superior test to the old mtDNA and microsatellite tests.
Results from the test are provided as a percentage "cattle score", but such is not defined and the meaning of the cattle score is left to one's own interpretation. Many interpret it to mean percent cattle genes (introgression) but this may or may not be correct. Relying on the sub-species composition test data (11) leaves many unanswered questions.
In 2022, a study published out of Texas A&M University, also employing whole genome snp genotyping, provided evidence for recent introgression in all bison including those animals previously thought to be free from hybridization with domestic cattle (12). Their detailed analyses using whole genome snp sequencing from nineteen modern and six historical bison, chosen to represent the major lineages of bison, identified and quantified signatures of recent nuclear introgression (within 200 years) in all bison. Although introgression regions varied greatly in size and frequency by sample and herd, both low and high coverage bison genomes provided evidence for domestic cattle introgression in all bison genomes tested.
After all these years ignoring the issue and indiscriminate breeding, it is not unreasonable to conclude that all bison have some level of cattle introgression as suggested by the 2022 Texas A&M publication (12). However, there are issues with these latest studies that bring at least some of the findings into question.
Although whole genomic snp genotyping is a powerful and even the preferred methodology tool, it is not without error, provides low information content, and is prone to ascertainment bias.
There was also a lack of any negative control in both introgression studies. In other words, all animals had evidence of introgression. Negative controls are essential for detection and correction of unmeasured confounding bias as well as detecting both suspected and unsuspected sources of spurious causal inference. The failure to incorporate any negative controls is a threat to the validity of the studies. Negative controls were available even if they needed to go to ancient bison before European or even humans inhabited North America (13). Negative controls were and are available. The lack of negative controls casts doubt on whether all these introgression markers are recent events.
Determining if the data is evidence of introgression or shared ancestral polymorphism is always problematic and the lack of controls compounds these issues. Are the observed patterns of shared alleles between species really the product of recent introgression, as opposed to non-contemporary processes such as ancient introgression or the incomplete lineage sorting of genes after speciation (14, 15).
The lack of negative controls likely accounted for the authors stating that their "putative cattle haplotypes" and "putative introgressed regions" have a "high likelihood of cattle origin" and "potential introgression" (emphasis added). Furthermore, the authors note that the introgression detected in their "historic" bison samples, allegedly considered to represent the last remaining wild bison populations, could not be explained (12). While they offer some explanations, they do not consider statistical error or that some alleles could be incorrectly defined as cattle markers.
Whether all bison have some evidence of cattle introgression or whether there are some genetically "pure" bison will likely be debated for decades to come. If we assume that all bison have some level of cattle introgression, do we continue to simply ignore it? Do we say it doesn't matter because all bison are introgressed? This unfortunately is the attitude taken by most of the industry. There are many bison with 5-10% (or more) cattle introgression and if we continue to indiscriminately sell and breed these animals we will continue to pollute the bison genome.
So, how do we interpret these data on cattle introgression?
My Interpretation (you may have another)

Plains Bison. Bison-type mtDNA, absent of all 25 microsatellite cattle markers, and 0% cattle score on Neogen/CBA introgression test.
We initially assembled our herd under a strict test and cull protocol employing mtDNA and nuclear microsatellite testing. We went directly through University of California-Davis for all our testing using the advanced microsatellite testing which looked for 25 introgression makers as opposed to the original 14. Any animal that had any evidence of introgression were culled which at the time accounted for about 30% of all bison we purchased. Thus we considered our herd to be free of cattle introgression.
Using the NeoGen/CBA test, two of our animals that previously tested clean had a 3% and 4% cattle score respectively. The remainder of the herd tested below 1%, most below 0.5%, and a few at 0.0%. The animal with a 3% and 4% cattle score and all their offspring were culled, but what to do or interpret those with less than 1%?
Biological science is not an exact science and is prone to bias and error. It is generally accepted within the biomedical community that as you increase sensitivity you decrease specificity. These recent introgression studies employed the "find the needle in the haystack" principle in which they essentially looked at every single strand of hay (without negative controls). Potential sources of error included, but were not limited to, statistical error, incorrectly defined recent cattle alleles, incomplete lineage sorting, errors in gene tree estimation, technical error such as systematic bias, cross-validation errors, and reliance on D-statistic which does not provide an unbiased estimation of the extent of introgression (16,17). These potential sources of error are compounded by the lack of negative controls.
So, capitalizing on the authors own language ("putative", aka assumed, "likelihood", and "potential introgression", I have adopted the following interpretation, definitions, and application:
- The percent "cattle score" is interpreted to mean the "likelihood" or "potential" of cattle introgression hereafter referred to as the probability of cattle introgression.
- Animals with greater than a 1% cattle score have a high likelihood of cattle introgression and are culled.
- Animals with less than 1% cattle score have a low likelihood of cattle introgression and are considered "pure" bison for all practical purposes.
Based on experience with the NeoGen test in generational testing, some of the cattle markers (snp's) used in testing appear to be heterologous (Figure 1 – i.e., the snp does not exist in both strands of the DNA double helix) and therefore could potentially be eliminated from the genome. What that means is that with selective breeding, some low likelihood animals could potentially be reverted back to the original bison genome.
What this means is that, even if Texas A&M is correct in suggesting that all bison are introgressed with cattle genes, by selective breeding of low likelihood animals, it may be possible to re-establish the original wild-type bison genome. That is what we are attempting to do.
Ozark Valley Bison Ranch LLC
Fox, AR
Literature Cited
1. Polziehn RO, Strobeck C, Sheraton J, Beech R. 1995. Bovine mtDNA Discovered in North American Bison Populations. Conservation Biology. 9:1638-43.
2. Dratch PA and PJP Gogan. 2010. Bison Conservation Initiative: Bison Conservation Genetics Workshop: report and recommendations. Natural Resource Report NPS/NRPC/BRMD/NRR—2010/257. National Park Service, Fort Collins, Colorado.
3. Hedrick PW. 2009. Conservation genetics and North American bison (Bison bison). The Journal of Heredity, 100(4): 411-420.
4. Boyd MM. 1908. Crossing Bison with Cattle. A Short Account of an Experiment in Crossing the American Bison with Domestic Cattle. American Breeders' Association. Vol, IV. Pp 324-331
5. Jones CJ. 1907. Breeding Cattelo. American Breeders' Association Annual Report, Vol. III. Page 161.
6. Dafoe JW. 1889. Domestication of the buffalo. Popular Science Monthly. 34:777-782.
7. Boyd MM. 1914. Crossing Bison and Cattle. J. Hered. 5:189–197.
8. Goodnight C. 1914. My experience with bison hybrids. J. Hered. 5:197–199.
9. Derr JN, Hedrick PW, Halbert ND, et al. 2012. Phenotypic effects of cattle mitochondrial DNA in American bison. Conservation Biology26:1130-1136.
10. Twyford AD and RA Ennos. 2012. Next-generation hybridization and introgression. Heredity 108:179-189.
11. Yang T, Miller M, Forgacs D, Derr J, Stothard P. 2020. Development of SNP-Based Genomic Tools for the Canadian Bison Industry: Parentage Verification and Subspecies Composition. Front. Genet. 11:585999.
12. Stroupe S, Forgacs D, Harris A, Derr JN, Davis BW. 2022. Genomic evaluation of hybridization in historic and modern North American Bison (Bison bison). Sci. Rep. 12:6397.
13. Froese D, Stiller M, Heintzman PD, Reyes AV, Zazula GD, et al. 2017. Fossil and genomic evidence constrains the timing of bison arrival in North America. Proc. Natl. Acad. Sci. U S A. 114: 3457–3462.
14. Pollard DA, Iyer VN, Moses AM, Eisen MB. 2006. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2:1634–1647.
15. Willyard A, Cronn R, Liston A. 2009. Reticulate evolution and incomplete lineage sorting among the ponderosa pines. Mol Phylogenet Evol. 52:498–511.
16. Pfeifer B, Kapan DD. 2019. Estimates of introgression as a function of pairwise distances. BMC Bioinformatics. 20:207.
17. Hamlin JAP, Hibbins MS, Moyle LC.. 2020. Assessing biological factors affecting postspeciation introgression. Evol Lett. 4:137–154.
